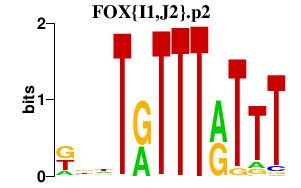
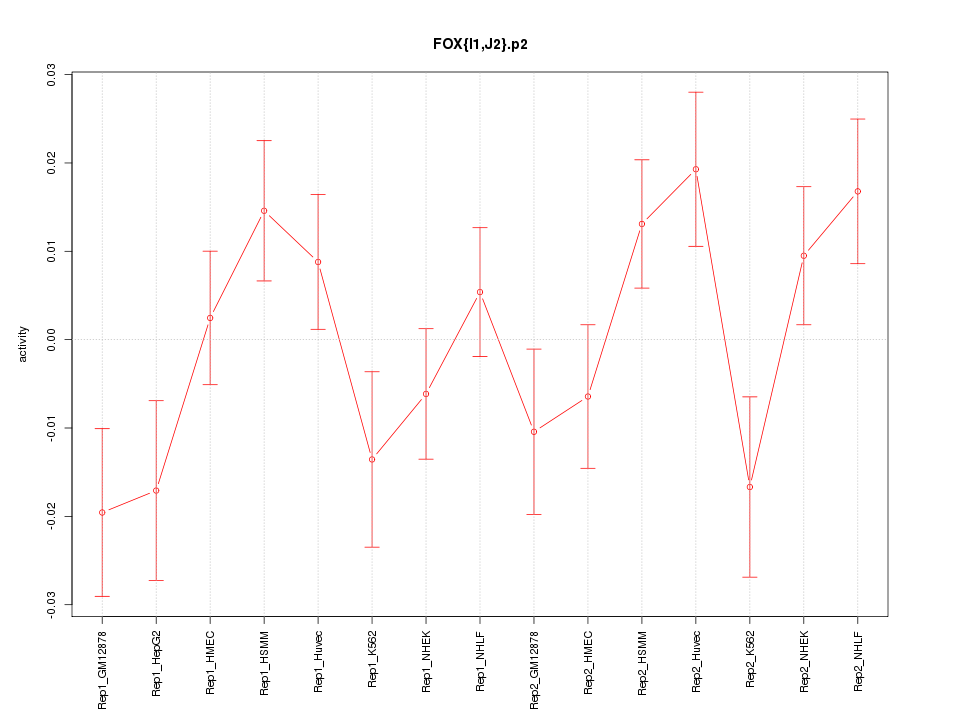
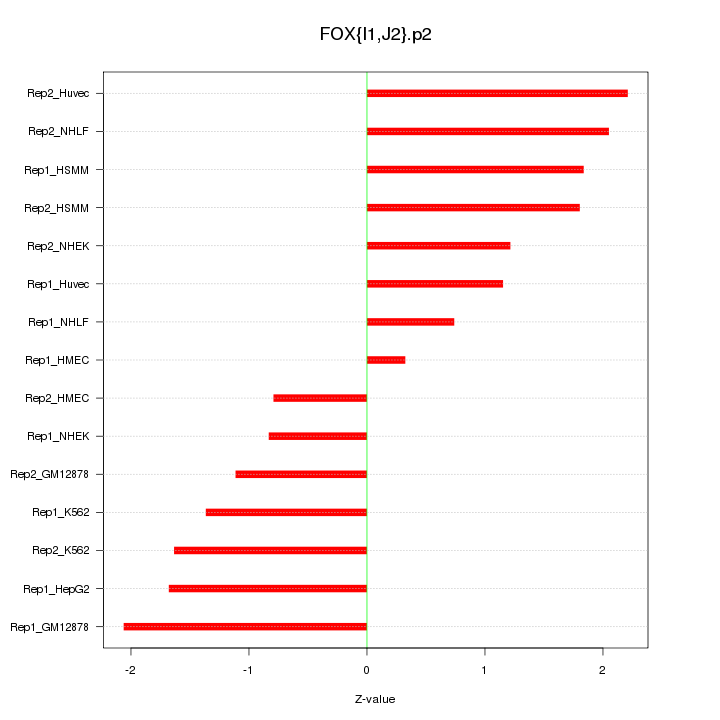
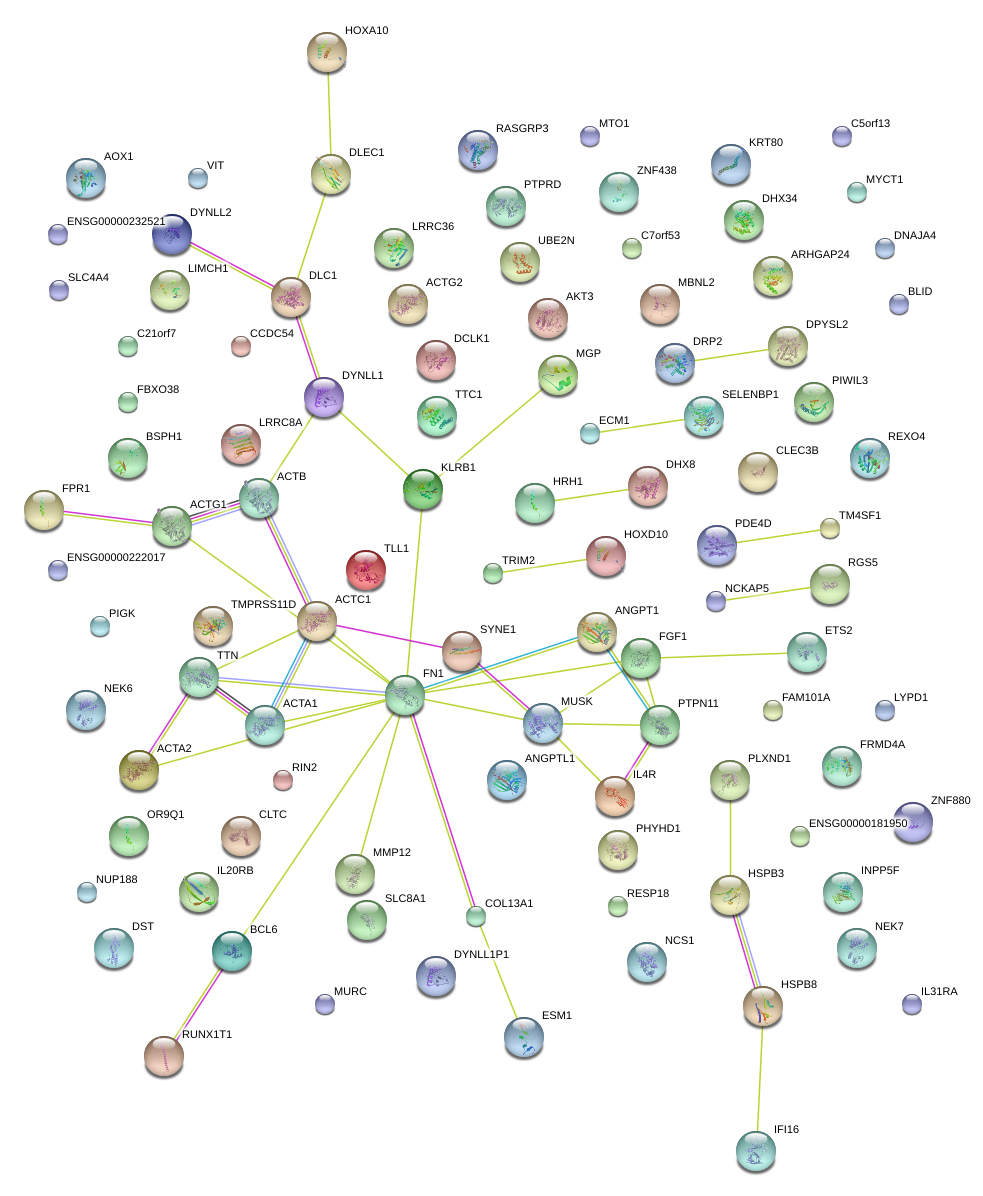
|
chr20_+_19818209
|
8.522
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr9_+_130719034
|
5.361
|
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr5_+_159368690
|
5.126
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr5_+_159368721
|
4.366
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr5_+_159368733
|
4.032
|
NM_003314
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr1_-_77457664
|
3.621
|
NM_005482
|
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K
|
|
chr2_-_133145539
|
3.564
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr4_+_167013859
|
3.353
|
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr2_+_201182187
|
3.237
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr17_+_55098280
|
3.223
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr5_+_147775652
|
3.041
|
|
FBXO38
|
F-box protein 38
|
|
chr12_+_111375382
|
2.822
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr4_+_86968069
|
2.725
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr3_-_130761967
|
2.649
|
|
PLXND1
|
plexin D1
|
|
chr11_+_57547928
|
2.634
|
NM_001005212
|
OR9Q1
|
olfactory receptor, family 9, subfamily Q, member 1
|
|
chr4_+_154397975
|
2.620
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr22_-_41366550
|
2.608
|
NM_001165877
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2
|
|
chr5_+_55182963
|
2.597
|
NM_139017
|
IL31RA
|
interleukin 31 receptor A
|
|
chr2_-_40593074
|
2.581
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr2_-_134042500
|
2.504
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr2_-_215998181
|
2.383
|
|
FN1
|
fibronectin 1
|
|
chr21_+_39117064
|
2.297
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr13_-_35327798
|
2.198
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr10_-_90702490
|
2.183
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr9_+_102380156
|
2.160
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr5_+_53787187
|
2.119
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr7_+_111908143
|
2.114
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr12_-_50871953
|
2.096
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr15_+_76345577
|
2.096
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr9_+_130722994
|
2.043
|
NM_001100876
NM_174933
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr2_+_36777406
|
2.031
|
|
VIT
|
vitrin
|
|
chr5_-_54317102
|
2.030
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr6_-_56615544
|
2.028
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr1_+_148747110
|
2.011
|
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr11_-_102250876
|
1.881
|
NM_002426
|
MMP12
|
matrix metallopeptidase 12 (macrophage elastase)
|
|
chr8_-_93181091
|
1.832
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_149611768
|
1.785
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr5_-_59100194
|
1.755
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr21_+_29439768
|
1.721
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr9_+_674193
|
1.716
|
|
|
|
|
chr10_+_71231649
|
1.711
|
NM_001130103
NM_080798
NM_080800
NM_080801
NM_080802
NM_080805
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr12_+_123339662
|
1.707
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr10_+_71231693
|
1.699
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr10_-_31328426
|
1.673
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr7_-_27186400
|
1.660
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr8_-_108579270
|
1.601
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr4_+_72321113
|
1.570
|
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr3_+_45042754
|
1.513
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr6_+_74228331
|
1.507
|
|
MTO1
|
mitochondrial translation optimization 1 homolog (S. cerevisiae)
|
|
chr22_+_44828338
|
1.500
|
|
|
|
|
chr2_+_36777336
|
1.485
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr19_-_56946911
|
1.436
|
NM_001193306
NM_002029
|
FPR1
|
formyl peptide receptor 1
|
|
chr2_-_219906142
|
1.396
|
NM_001007089
|
RESP18
|
regulated endocrine-specific protein 18 homolog (rat)
|
|
chr2_-_179380301
|
1.387
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr19_-_53187151
|
1.384
|
NM_001128326
|
BSPH1
|
binder of sperm protein homolog 1
|
|
chr5_-_142057692
|
1.377
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr1_-_242073094
|
1.371
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr6_+_153060722
|
1.359
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr9_+_112470871
|
1.357
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr2_+_33514896
|
1.350
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr9_+_132002692
|
1.346
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr4_-_68432293
|
1.345
|
NM_004262
|
TMPRSS11D
|
transmembrane protease, serine 11D
|
|
chr11_-_121492010
|
1.342
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr9_-_8723893
|
1.332
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr8_-_13416554
|
1.315
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr16_+_65938753
|
1.293
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr22_-_23500682
|
1.286
|
NM_001008496
|
PIWIL3
|
piwi-like 3 (Drosophila)
|
|
chr8_-_93176618
|
1.283
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_-_14929991
|
1.278
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr3_+_108578877
|
1.278
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr4_+_41309482
|
1.263
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr10_-_14090459
|
1.260
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr12_+_118100977
|
1.246
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr6_-_152681171
|
1.243
|
NM_015293
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr9_+_126094067
|
1.241
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr13_+_20081940
|
1.224
|
|
|
|
|
chr3_-_150569574
|
1.223
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr3_-_188936941
|
1.223
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr3_+_11242666
|
1.220
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr13_+_96726197
|
1.206
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chrX_+_100361588
|
1.204
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr3_+_138159396
|
1.194
|
NM_144717
|
IL20RB
|
interleukin 20 receptor beta
|
|
chr19_+_57564981
|
1.193
|
NM_001145434
|
ZNF880
|
zinc finger protein 880
|
|
chr1_-_161439487
|
1.178
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr16_+_27248806
|
1.171
|
|
IL4R
|
interleukin 4 receptor
|
|
chr5_-_111340421
|
1.167
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_157246305
|
1.155
|
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr20_+_19863716
|
1.142
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr1_-_177106702
|
1.134
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr15_+_65205107
|
1.134
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr1_-_171286634
|
1.132
|
NM_005092
|
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18
|
|
chr7_+_150442652
|
1.130
|
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr15_+_37330176
|
1.121
|
NM_207445
|
C15orf54
|
chromosome 15 open reading frame 54
|
|
chr1_+_196148241
|
1.121
|
NM_001014434
|
LHX9
|
LIM homeobox 9
|
|
chr7_-_112513379
|
1.113
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr2_-_183095308
|
1.101
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr18_+_53863865
|
1.097
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_+_156229686
|
1.095
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr13_-_32678142
|
1.093
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_-_119768430
|
1.091
|
|
|
|
|
chr7_+_17349425
|
1.089
|
|
AHR
|
aryl hydrocarbon receptor
|
|
chrX_-_19598912
|
1.088
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr7_-_27125738
|
1.087
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr5_-_36337756
|
1.081
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr5_+_68702287
|
1.073
|
NM_002873
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chrX_+_135106578
|
1.068
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr3_+_46423682
|
1.067
|
NM_003965
NM_001130910
|
CCRL2
|
chemokine (C-C motif) receptor-like 2
|
|
chr2_-_99412694
|
1.066
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr3_+_137224266
|
1.065
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr1_-_166965034
|
1.058
|
NM_001937
|
DPT
|
dermatopontin
|
|
chr19_+_9157269
|
1.044
|
NM_175883
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2
|
|
chr2_-_72228427
|
1.037
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr1_+_22107137
|
1.035
|
|
RPL21
|
ribosomal protein L21
|
|
chr2_+_151922350
|
1.032
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr18_+_7936885
|
1.015
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr12_-_90100733
|
1.012
|
NM_001920
|
DCN
|
decorin
|
|
chr12_+_26017954
|
0.996
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr12_-_120220493
|
0.979
|
NM_006549
NM_153499
NM_153500
NM_172214
NM_172215
NM_172216
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr5_+_81636921
|
0.976
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr5_-_125956996
|
0.976
|
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr8_-_52884456
|
0.973
|
NM_144651
|
PXDNL
|
peroxidasin homolog (Drosophila)-like
|
|
chr12_-_90100679
|
0.971
|
|
DCN
|
decorin
|
|
chr8_+_32698892
|
0.961
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr13_+_75092570
|
0.961
|
NM_005358
|
LMO7
|
LIM domain 7
|
|
chr9_-_16717887
|
0.960
|
|
BNC2
|
basonuclin 2
|
|
chr6_+_134800533
|
0.958
|
|
LOC154092
|
hypothetical LOC154092
|
|
chr12_+_79362256
|
0.949
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr11_-_8695974
|
0.947
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr12_+_47584159
|
0.946
|
NM_033124
|
CCDC65
|
coiled-coil domain containing 65
|
|
chr8_+_120148604
|
0.941
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr16_+_31036474
|
0.933
|
NM_032188
NM_182958
|
MYST1
|
MYST histone acetyltransferase 1
|
|
chr18_-_44723116
|
0.933
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr1_-_110735158
|
0.929
|
NM_004696
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr1_+_143807748
|
0.926
|
NM_004892
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr1_+_84540921
|
0.920
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr14_+_57964462
|
0.917
|
NM_014749
|
KIAA0586
|
KIAA0586
|
|
chr4_-_186815116
|
0.911
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_+_38439399
|
0.910
|
NM_182798
NM_182799
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr2_-_225519974
|
0.903
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr2_-_215951161
|
0.901
|
|
FN1
|
fibronectin 1
|
|
chr9_-_4290028
|
0.896
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr11_-_128279670
|
0.894
|
|
C11orf45
|
chromosome 11 open reading frame 45
|
|
chr22_+_40372957
|
0.891
|
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6
|
|
chr1_-_157950898
|
0.881
|
NM_000567
|
CRP
|
C-reactive protein, pentraxin-related
|
|
chr8_-_93177057
|
0.879
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_-_2814410
|
0.874
|
NM_031474
|
NRIP2
|
nuclear receptor interacting protein 2
|
|
chr1_+_84382529
|
0.872
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr11_-_101292462
|
0.869
|
NM_178127
|
ANGPTL5
|
angiopoietin-like 5
|
|
chr1_+_66570363
|
0.861
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr8_+_75066117
|
0.857
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr19_-_53305825
|
0.855
|
NM_001159322
NM_001159323
NM_003706
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent)
|
|
chr19_-_59258846
|
0.847
|
NM_198481
|
VSTM1
|
V-set and transmembrane domain containing 1
|
|
chr13_-_109600709
|
0.844
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr1_+_66570683
|
0.844
|
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr13_+_22653059
|
0.840
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr8_-_18710575
|
0.838
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr6_-_138935359
|
0.837
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr3_+_150675117
|
0.836
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr6_+_121798443
|
0.832
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr2_-_175170674
|
0.822
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr10_+_94584449
|
0.819
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr2_-_219404755
|
0.813
|
NM_017431
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit
|
|
chr6_+_151688327
|
0.812
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr12_-_120219941
|
0.812
|
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr3_+_150675169
|
0.811
|
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr1_-_149245503
|
0.795
|
NM_001163259
NM_001163260
NM_001040217
|
FAM63A
|
family with sequence similarity 63, member A
|
|
chr10_-_116276560
|
0.795
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr5_+_149977398
|
0.790
|
NM_001166209
|
SYNPO
|
synaptopodin
|
|
chr2_+_173394560
|
0.786
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr11_-_30910074
|
0.786
|
NM_020869
|
DCDC5
|
doublecortin domain containing 5
|
|
chr4_-_186814852
|
0.785
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr17_+_53587492
|
0.784
|
NM_012374
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1
|
|
chrX_-_43717864
|
0.783
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr8_+_70567581
|
0.783
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr3_-_170970296
|
0.782
|
NM_032487
|
ARPM1
|
actin related protein M1
|
|
chr21_-_42219849
|
0.771
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr5_-_59100091
|
0.768
|
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_87687276
|
0.767
|
|
SRI
|
sorcin
|
|
chr5_+_159546951
|
0.764
|
NM_001130958
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr17_+_7530113
|
0.761
|
NM_001143990
|
WRAP53
|
WD repeat containing, antisense to TP53
|
|
chr1_-_168310232
|
0.760
|
NM_014970
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr9_+_119506274
|
0.756
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr17_+_33867169
|
0.753
|
NM_020876
|
ARHGAP23
|
Rho GTPase activating protein 23
|
|
chr3_-_102194938
|
0.751
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr7_+_136203938
|
0.750
|
NM_001006627
NM_001006630
|
CHRM2
|
cholinergic receptor, muscarinic 2
|
|
chr3_-_11598829
|
0.748
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr9_-_4289915
|
0.746
|
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr16_+_67698943
|
0.746
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr9_-_116608113
|
0.745
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr18_+_30544193
|
0.741
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_-_54119159
|
0.741
|
NM_032622
|
LNX1
|
ligand of numb-protein X 1
|
|
chr2_+_108571336
|
0.739
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr9_+_5500544
|
0.739
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr21_-_36774257
|
0.734
|
NM_001146079
NM_144492
|
CLDN14
|
claudin 14
|
|
chr4_-_130233855
|
0.729
|
NM_144643
|
SCLT1
|
sodium channel and clathrin linker 1
|
|
chr14_+_78815406
|
0.728
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr9_-_116920232
|
0.725
|
NM_002160
|
TNC
|
tenascin C
|
|
chr13_-_77391525
|
0.723
|
|
|
|
|
chr4_-_54119123
|
0.722
|
|
LNX1
|
ligand of numb-protein X 1
|
|
chr8_-_93176819
|
0.720
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|